GenoScreen services
Sanger sequencing - The low-throughput genomics solution
Sanger sequencing is a well-proven, low-throughput analytical solution that is well suited to fragments of all sizes (100-1200 bp read length). GenoScreen’s Sanger sequencing services can be adapted to suit all types of project, regardless the size, delay time or frequency.
This email address is being protected from spambots. You need JavaScript enabled to view it.
Sanger sequencing applications
Despite the emergence of new generation technologies (NGS), Sanger sequencing remains a reference method in many fields thanks to its reliability and accuracy.
GenoScreen offer
Flexible and available, our teams carry out sequencing from sample preparation to data analysis. See our documentation below to find out more about our services and to prepare your sequencing request form.
Premium range
The Premium offers correspond to a complete and personalised analysis of samples:
- Adaptation of the protocol to GC-rich sequences
- Supply of universal primers
- Sequence reading up to 1200 bp
- Results sent within 24 to 48 hours
The Premium range gives you access to our hotline, by phone and email: expert scientific support for sequence profile analysis.
Workflow tailored to your needs
All our offers are open to different sample formats:
- tubes
- strips
- plates, complete or not
Our One-Shot Plus and Optimized offers include additional information:
- Signal optimisation
- Reduction of any backgroun noise
- Assignment of indeterminate bases
- A specific protocol for mutation detection
Additional analyses
GenoScreen offers a range of services to complement sequencing:
Economic range
All the quality and responsiveness of GenoScreen at a low price!
Using your own PCR product + primer mix or even the sequencing reaction you have produced, we can take your samples to a more advanced stage for an optimised price.
GenoScreen advantages
- Flexible sequencing offer that adapts to your needs
-
Personalised monitoring of your projects
-
Simple ordering system
-
Monthly invoicing or prepaid account
-
Free transport of your samples by a GenoScreen-approved operator
-
Optimised protection of samples in Genobox

Metagenomics: powerful analyses of microbial communities
Molecular metagenomics is used to analyze a community of organisms as a whole; as such, it provides an exhaustive assessment of a complex ecological niche. It is a powerful, innovative approach for characterizing the genes and thus the biological functions in a given microbiome.
Metagenomics has many advantages:
- Removal of the bias associated with PCR approaches
- Culture-free analysis and direct sequencing of microorganisms
- The functional description of a whole ecosystem
Applications
Metagenomics and human health
Metagenomics and the environment
In credit to its very detailed analysis of microbial communities present in the natural environment, metagenomics can be applied to the analysis of water quality, the development of biofuels and agrochemical products, soil remediation and the development of new practices in agriculture and animal husbandry.
GenoScreen - Services provided
GenoScreen's assets
- A dedicated team of expert scientists for each project.
- Assistance with developing the experimental plan and the sequencing strategy.
- Massive whole metagenome sequencing capacity.
- Exclusive bioinformatics tools.
- Cutting-edge scientific expertise for data exploitation (specially in the field of microbiology).
Targeted sequencing - An in-depth analysis of regions of interest
Targeted sequencing allows specific analysis of targeted genes (or gene regions) on many samples in parallel. With NGS sequencing on MiSeq®, NextSeq500® or Hiseq® platforms, multiple genes can be evaluated with a smaller data volume, which is easier to manage and analyse than WGS datas.
Applications
Sequencing or targeted resequencing can achieve very specific objectives:
- Screening of genomic regions of interest.
- Complete coverage of coding regions, with a limited volume of genomic data.
- Analysis of microbial genes.
GenoScreen - Services provided
Amplicon sequencing
A number of the target regions are amplified by PCR, using the initially defined specific primers. The amplicons obtained are then sequenced by NGS, following the addition of sequencing adaptors. Multiplexing approaches enable us to sequence several amplicons from different samples in parallel.
The Metabiote® solution
Metabiote® is a dedicated, innovative, optimized, standardized solution developed by GenoScreen. It integrates targeted metagenomic analysis of microbial communities.
- A library preparation methodology that limits between-sample bias.
- A diverse range of targets:
• bacterial targets: 16S rDNA
• fungal targets: 18S, ITS1 and/or ITS2 rDNA - A bioinformatics pipeline that has been optimized and automated for high-precision analyses, easy comparisons and statistical analyses, and a high degree of responsiveness.
Exome sequencing
This type of sequencing is essentially used to:
- Screen for mutations in genes involved in regulatory pathways
- Screen for mutations (SNPs, indels, etc.) associated with a particular disease (cancer, coagulopathies, etc.).
We use the NimbleGen SeqCap EZ protocol (Roche) for exome sequencing.
Personalized sequencing
After custom probe design, your regions of interest are enriched prior to sequencing which simplifies the subsequent data analysis. This protocol can be set up for any organism with an available reference genome.
GenoScreen's assets
- A dedicated team of scientists for each project.
- Comprehensive support with the elaboration of an experimental plan and project implementation.
- Assistance with data handling and interpretation of the results.
Ready-to-load services - Plugged into our sequencing platform
For a cost effective and responsive offer, GenoScreen proposes "Ready to Load" sequencing services for a wide range of libraries: a real research accelerator giving the opportunity to perform on last generation technical platform with advanced expertise.
Applications
"Ready-to-load" applications are available for mainly all of GenoScreen’s sequencing services:
- De novo sequencing or re-sequencing.
- RNA-Seq.
- Targeted sequencing.
- Metagenomics.
GenoScreen - Services provided
"Ready-to-load" sequencing is performed on latest-generation technical facilities: Illumina HiSeq®2500, MiSeq®, NextSeq®500 and HiSeq®4000 systems in paired-end or single-end runs of 50 to 250 bp.
GenoScreen performs quality control on the libraries with bioAnalyzer and fluorimetric assays. GenoScreen can also ensure the pooling of the sequencing libraries, if required.
We accept different types of librairies:
- TruSeq.
- Nextera(-XT).
- Amplicons.
- RADseq or ddRADseq with index and Illumina adaptors.
For the other types of librairies, please contact us.
The results
- Raw sequences (FastQ files) and quality control results after demultiplexing.
- Quality reports, including the number of reads obtained, the number of reads after application of quality filters, and the coverage obtained.
- The results are transferred to a secure FTP server or an external hard disk.
GenoScreen's assets
- Cutting-edge technical facilities.
- A top-of-the-range service at a keen price.
- Responsiveness.
Transcriptomics - Analyzing gene expression
High-throughput sequencing of the transcriptome (RNA-Seq) has revolutionized the quantitative and qualitative analysis of prokaryotic and eukaryotic organisms.
RNA-Seq significatively enables differential expression analyses:
- Measurement of the relative abundance of transcripts.
- Identification of the genes expressed differentially in various groups (tissues, treatments, etc.)
Applications
Transcriptome sequencing provides a complete, objective analysis of messenger RNAs:
- Differential expression: assessment of variations in the expression of several transcriptomes.
- Annotation of a transcriptome against a reference database.
- Identification of all specific sequences of the studied transcriptome.
- SNP analysis.
GenoScreen - Services provided
Two methods are used to enrich the RNA fraction of interest (mRNA) prior to cDNA synthesis and library preparation:
- Poly(A) capture (for eukaryotic total RNA).
- RNA depletion (via capture with complementary probes) and mRNA enrichment (for prokaryotic or eukaryotic total RNA).
Most of our RNA-Seq libraries are sequenced on Illumina® HiSeq®4000 systems.
GenoScreen's assets
- A dedicated team of scientists for each project.
- Assistance with elaborating the experimental plan and sequencing strategy.
- RNA extraction, quantification, purification, and quality control.
- Assistance with data management and interpretation of the results.
- Analysis of differential expression in the absence of a reference genome.
Whole-genome sequencing - A high-precision method for characterizing the genome
The latest sequencing technologies make it possible to analyze the full primary structure of an isolated organism’s genome, regardless of, whether the latter is a eukaryote, a prokaryote or a virus.
An analysis of whole-genome sequencing data provides a precised description of any type of organism and thus addresses a very broad range of research questions.
Applications
Whole-genome sequencing addresses important research questions in many sectors (healthcare, nutrition, agrifood, environment, etc.):
- Description of an organism’s genes and how they are organized.
- Quantification of diagnostic or phenotypic biomarkers.
- Screening for susceptibility/adaptability/resistance genes.
GenoScreen - Services provided
As specialists in whole-genome sequencing (WGS), our expert teams are able to build you a custom solution.
Our precise, fast solutions combine NGS and Sanger techniques and are implemented in cutting-edge technical facilities.
Our bioinformatics solutions can, then deliver in-depth processing and analysis of your WGS data (assembly, annotation, etc.).
- Genome structure and mapping of isolated organisms.
- Comparative genomics: identification of SNPs, indels and/or major structural rearrangements.
- Multilocus sequence typing (MLST).
GenoScreen's assets
- We understand your needs and provide personalized support.
- Exclusive bioinformatics analyses.
- Significant expertise in sequencing microorganisms (bacteria, fungi, yeasts, algae, etc.).
- Fast solution.
MLST/MLVA, microbial typing/tracing solutions
GenoScreen proposes you the opportunity to characterize more accurately your microorganisms (subspecies or strain characterization) using standardized and validated molecular biology methods such as MLST or MLVA (including MIRU-VNTR).
MultiLocus Sequencing Typing (MLST)
The MLST by GenoScreen
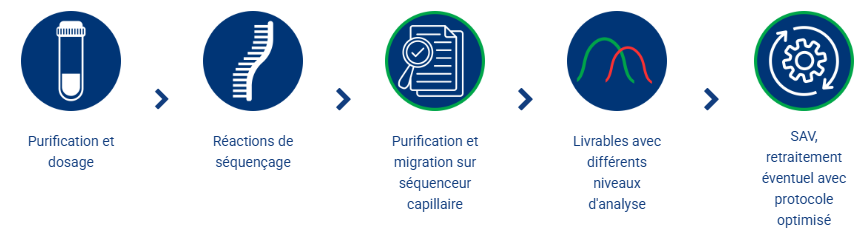
GenoScreen's MLST service offer is a complete package of services realized from extracted DNA or thermolysate. This includes the PCRs realization, the sequencing on a capillary sequencer, the sequences analysis and the strain assignment..
| Règne / Kingdom | Classe / Class | Ordre / Order | Espèce / Species |
|---|---|---|---|
| Animalia | Myxozoa | Multivalvulida | Kudoa septempunctata |
| Animalia | Rhabditophora | Plagiorchiida | Clonorchis sinensis |
| Bacteria | Actinobacteria | Actinomycetales | Corynebacterium diphtheriae |
| Bacteria | Actinobacteria | Actinomycetales | Cutibacterium acnes |
| Bacteria | Actinobacteria | Actinomycetales | Mycobacterium abscessus complex |
| Bacteria | Actinobacteria | Actinomycetales | Mycobacterium spp. |
| Bacteria | Actinobacteria | Actinomycetales | Rhodococcus equi |
| Bacteria | Actinobacteria | Actinomycetales | Streptomyces spp. |
| Bacteria | Alpha Proteobacteria | Rhizobiales | Bartonella bacilliformis |
| Bacteria | Alpha Proteobacteria | Rhizobiales | Bartonella henselae |
| Bacteria | Alpha Proteobacteria | Rhizobiales | Bartonella washoensis |
| Bacteria | Alpha Proteobacteria | Rhizobiales | Brucella spp. |
| Bacteria | Alpha Proteobacteria | Rhizobiales | Candidatus Liberibacter solanacearum |
| Bacteria | Alpha Proteobacteria | Rhizobiales | Sinorhizobium spp. |
| Bacteria | Alpha Proteobacteria | Rickettsiales | Anaplasma phagocytophilum |
| Bacteria | Alpha Proteobacteria | Rickettsiales | Orientia tsutsugamushi |
| Bacteria | Alpha Proteobacteria | Rickettsiales | Wolbachia spp. |
| Bacteria | Bacilli | Bacillales | Bacillus cereus |
| Bacteria | Bacilli | Bacillales | Bacillus licheniformis |
| Bacteria | Bacilli | Bacillales | Bacillus subtilis |
| Bacteria | Bacilli | Bacillales | Macrococcus canis |
| Bacteria | Bacilli | Bacillales | Macrococcus caseolyticus |
| Bacteria | Bacilli | Bacillales | Paenibacillus larvae |
| Bacteria | Bacilli | Bacillales | Staphylococcus aureus |
| Bacteria | Bacilli | Bacillales | Staphylococcus epidermidis |
| Bacteria | Bacilli | Bacillales | Staphylococcus haemolyticus |
| Bacteria | Bacilli | Bacillales | Staphylococcus hominis |
| Bacteria | Bacilli | Bacillales | Staphylococcus pseudintermedius |
| Bacteria | Bacilli | Lactobacillales | Carnobacterium maltaromaticum |
| Bacteria | Bacilli | Lactobacillales | Enterococcus faecalis |
| Bacteria | Bacilli | Lactobacillales | Enterococcus faecium |
| Bacteria | Bacilli | Lactobacillales | Lactobacillus salivarius |
| Bacteria | Bacilli | Lactobacillales | Melissococcus plutonius |
| Bacteria | Bacilli | Lactobacillales | Pediococcus pentosaceus |
| Bacteria | Bacilli | Lactobacillales | Streptococcus agalactiae |
| Bacteria | Bacilli | Lactobacillales | Streptococcus anginosus |
| Bacteria | Bacilli | Lactobacillales | Streptococcus bovis complex |
| Bacteria | Bacilli | Lactobacillales | Streptococcus canis |
| Bacteria | Bacilli | Lactobacillales | Streptococcus dysgalactiae |
| Bacteria | Bacilli | Lactobacillales | Streptococcus equinus complex |
| Bacteria | Bacilli | Lactobacillales | Streptococcus faecalis |
| Bacteria | Bacilli | Lactobacillales | Streptococcus gallolyticus |
| Bacteria | Bacilli | Lactobacillales | Streptococcus gordonii |
| Bacteria | Bacilli | Lactobacillales | Streptococcus mitis |
| Bacteria | Bacilli | Lactobacillales | Streptococcus mutans |
| Bacteria | Bacilli | Lactobacillales | Streptococcus oralis |
| Bacteria | Bacilli | Lactobacillales | Streptococcus pneumoniae |
| Bacteria | Bacilli | Lactobacillales | Streptococcus pyogenes |
| Bacteria | Bacilli | Lactobacillales | Streptococcus salivarius |
| Bacteria | Bacilli | Lactobacillales | Streptococcus suis |
| Bacteria | Bacilli | Lactobacillales | Streptococcus thermophilus |
| Bacteria | Bacilli | Lactobacillales | Streptococcus uberis |
| Bacteria | Bacilli | Lactobacillales | Streptococcus viridans |
| Bacteria | Bacilli | Lactobacillales | Streptococcus zooepidemicus |
| Bacteria | Bacteroidetes | Porphyromonadaceae | Porphyromonas gingivalis |
| Bacteria | Beta Proteobacteria | Burkholderiales | Achromobacter |
| Bacteria | Beta Proteobacteria | Burkholderiales | Bordetella spp. |
| Bacteria | Beta Proteobacteria | Burkholderiales | Burkholderia cepacia complex |
| Bacteria | Beta Proteobacteria | Burkholderiales | Burkholderia pseudomallei |
| Bacteria | Beta Proteobacteria | Burkholderiales | Taylorella spp. |
| Bacteria | Beta Proteobacteria | Neisseriales | Neisseria spp. |
| Bacteria | Chlamydiae | Chlamydiales | Chlamydiales spp. |
| Bacteria | Clostridia | Clostridiales | Clostridioides difficile |
| Bacteria | Clostridia | Clostridiales | Clostridium botulinum |
| Bacteria | Clostridia | Clostridiales | Clostridium septicum |
| Bacteria | Epsilon Proteobacteria | Campylobacterales | Arcobacter spp. |
| Bacteria | Epsilon Proteobacteria | Campylobacterales | Campylobacter spp. |
| Bacteria | Epsilon Proteobacteria | Campylobacterales | Helicobacter cinaedi |
| Bacteria | Epsilon Proteobacteria | Campylobacterales | Helicobacter pylori |
| Bacteria | Epsilon Proteobacteria | Campylobacterales | Helicobacter suis |
| Bacteria | Flavobacteria | Flavobacteriales | Flavobacterium psychrophilum |
| Bacteria | Flavobacteria | Flavobacteriales | Ornithobacterium rhinotracheale |
| Bacteria | Flavobacteria | Flavobacteriales | Riemerella anatipestifer |
| Bacteria | Flavobacteria | Flavobacteriales | Tenacibaculum spp. |
| Bacteria | Gamma Proteobacteria | Aeromonadales | Aeromonas spp. |
| Bacteria | Gamma Proteobacteria | Cardiobacteriales | Dichelobacter nodosus |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Citrobacter freundii |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Cronobacter spp. |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Edwardsiella spp. |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Enterobacter cloacae |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Escherichia spp. |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Klebsiella aerogenes |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | klebsiella michiganensis |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Klebsiella oxytoca |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Salmonella spp. |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Yersinia pseudotuberculosis |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Yersinia ruckeri |
| Bacteria | Gamma Proteobacteria | Enterobacteriales | Yersinia spp. |
| Bacteria | Gamma Proteobacteria | Pasteurellales | Gallibacterium anatis |
| Bacteria | Gamma Proteobacteria | Pasteurellales | Haemophilus influenzae |
| Bacteria | Gamma Proteobacteria | Pasteurellales | Haemophilus parasuis |
| Bacteria | Gamma Proteobacteria | Pasteurellales | Mannheimia haemolytica |
| Bacteria | Gamma Proteobacteria | Pasteurellales | Pasteurella multocida |
| Bacteria | Gamma Proteobacteria | Pseudomonadales | Acinetobacter baumannii |
| Bacteria | Gamma Proteobacteria | Pseudomonadales | Moraxella catarrhalis |
| Bacteria | Gamma Proteobacteria | Pseudomonadales | Pseudomonas aeruginosa |
| Bacteria | Gamma Proteobacteria | Pseudomonadales | Pseudomonas fluorescens |
| Bacteria | Gamma Proteobacteria | Pseudomonadales | Pseudomonas putida |
| Bacteria | Gamma Proteobacteria | Thiotrichales | Piscirickettsia salmonis |
| Bacteria | Gamma Proteobacteria | Vibrionales | Photobacterium damselae |
| Bacteria | Gamma Proteobacteria | Vibrionales | Vibrio cholerae |
| Bacteria | Gamma Proteobacteria | Vibrionales | Vibrio parahaemolyticus |
| Bacteria | Gamma Proteobacteria | Vibrionales | Vibrio spp. |
| Bacteria | Gamma Proteobacteria | Vibrionales | Vibrio tapetis |
| Bacteria | Gamma Proteobacteria | Vibrionales | Vibrio vulnificus |
| Bacteria | Gamma Proteobacteria | Xanthomonadales | Stenotrophomonas maltophilia |
| Bacteria | Gamma Proteobacteria | Xanthomonadales | Xylella fastidiosa |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma agalactiae |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma bovis |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma flocculare |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma hominis |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma hyopneumoniae |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma hyorhinis |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma iowae |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma pneumoniae |
| Bacteria | Mollicutes | Mycoplasmatales | Mycoplasma synoviae |
| Bacteria | Mollicutes | Mycoplasmatales | Ureaplasma spp. |
| Bacteria | Spirochaetes | Brachyspirales | Brachyspira spp. |
| Bacteria | Spirochaetes | Leptospirales | Leptospira spp. |
| Bacteria | Spirochaetes | Spirochaetales | Borrelia spp. |
| Bacteria | Spirochaetes | Spirochaetales | Treponema pallidum subsp. pallidum |
| Chromista | Oomycota | Saprolegniales | Saprolegnia parasitica |
| Excavata | Parabasalia | Trichomonadida | Trichomonas vaginalis |
| Fungi | Blastocystae | Blastocystida | Blastocystis sp. |
| Fungi | Eurotiomycetes | Eurotiales | Aspergillus fumigatus |
| Fungi | Saccharomycetes | Saccharomycetales | Candida albicans |
| Fungi | Saccharomycetes | Saccharomycetales | Candida glabrata |
| Fungi | Saccharomycetes | Saccharomycetales | Candida krusei |
| Fungi | Saccharomycetes | Saccharomycetales | Candida tropicalis |
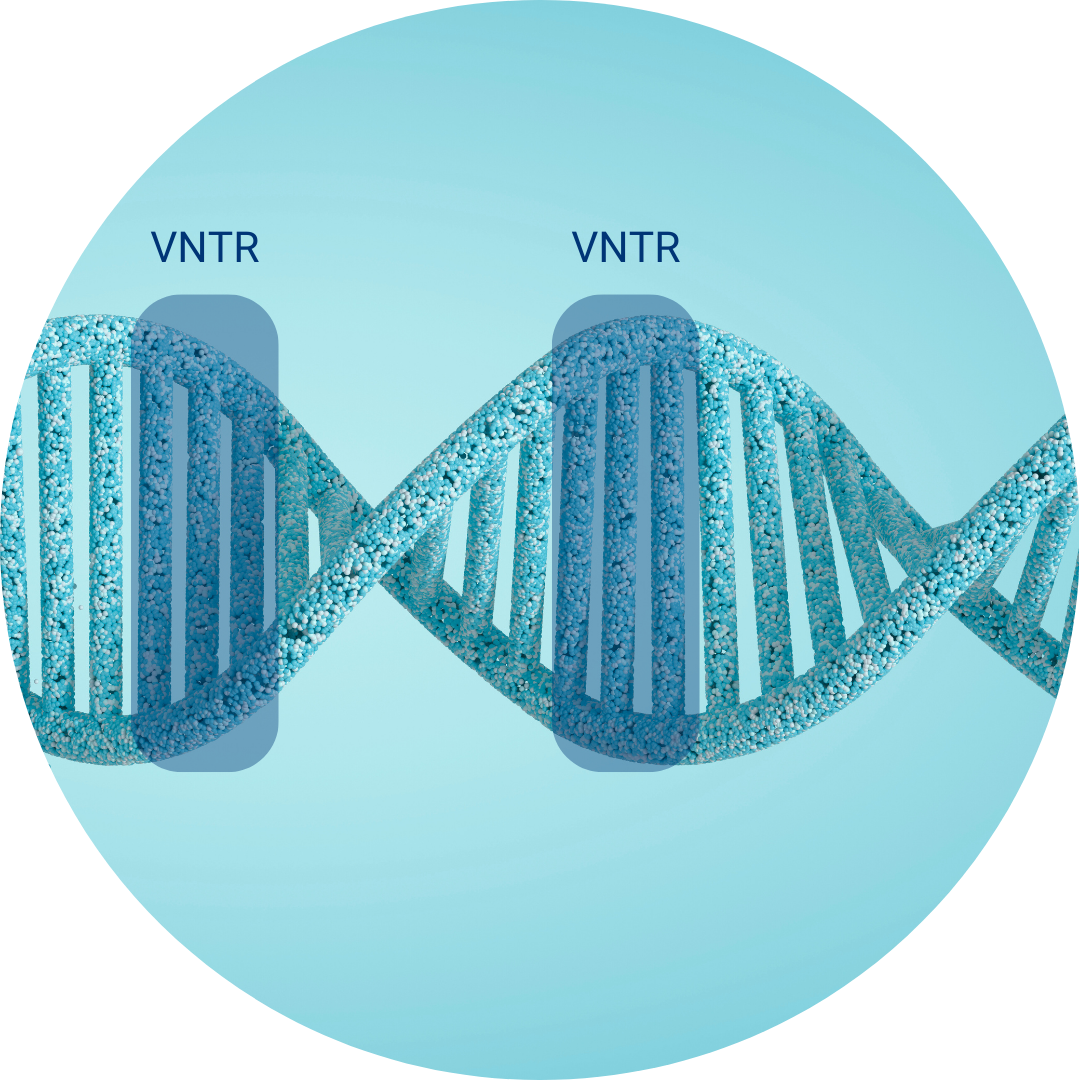
Multiple Loci VNTR Analysis (MLVA)
Le MLVA par GenoScreen
GenoScreen's MLVA service offer is a complete package of services realized from extracted DNA or thermolysate. This includes the PCRs realization, the electrophoresis on a capillary sequencer, the data analysis and the strain assignment.
The analysis can be carried out on the basis of a personalised typing scheme tailored to your needs, in line with the data available for your species.
| Règne / Kingdom | Classe / Class | Ordre / Order | Espèce / Species |
|---|---|---|---|
| Bacteria | Actinobacteria | Actinomycetales | Mycobacterium tuberculosis |
| Bacteria | Actinobacteria | Actinomycetales | Mycobacterium bovis |
| Bacteria | Actinobacteria | Actinomycetales | Mycobacterium avium |
| Bacteria | Gamma Proteobacteria | Thiotrichales | Francisella tularensis |
| Fungi | Saccharomycetes | Saccharomycetales | Saccharomyces cerevisiae |